FOR IMMEDIATE RELEASE

Students Elizabeth Honsaker, Daniel Pressman, Felicity Grady, Elizabeth Schramm, Catherine
Simpson watch as Dr. Tagide deCarvalho explains how to prepare samples for the lab’s
electron microscope. Dr. Steve Heninger (far right) observes the process. Not shown
is ACM Professor Michele Barmoy.
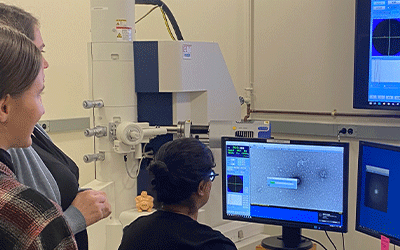
Student Elizabeth Honsaker and Felicity Grady view phages using UMBC's Transmission
Electron Microscope, guided by Dr. Tagide deCarvalho.
CUMBERLAND, Md. (Dec. 15, 2023)– Allegany College of Maryland (ACM) student researchers
Felicity Grady, Elizabeth Honsaker, Daniel Pressman, Elizabeth Schramm and Catherine
Simpson traveled to UMBC’s Keith R. Porter Imaging Facility on November 15. The five
students brought a cooler of four fresh bacteriophage (phage) lysates samples and
were joined by ACM Professors Michèle Barmoy and Steve Heninger.
Students used the Porter Imaging Facility’s Transmission Electron Microscope (TEM)
to get their first glimpse of their phages.
Tagide deCarvalho, who manages UMBC’s Porter Imaging Facility, showed the students
how to prepare samples for the lab’s TEM, a Hitachi HT7800 120 kV TEM with an AMT
Nanosprint15 B digital camera. Students mounted their phages on small copper grids
and coated them with Uranium acetate. When placed in the TEM, a beam of electrons
was transmitted through their phages. deCavralho and the student located intact phages
and photographed them using the TEM’s digital camera. Digital files of the images
were shared with the students and became part of their research.
Four of the students extracted enough DNA from their phages this semester to have
them sequenced by the University of Pittsburgh over winter break.
In the spring semester, students will annotate (identify the genes and their functions)
the sequenced DNA using bioinformatics (the use of computer software and scientific
methodology to understand data).
- Those results are submitted to the National Center for Biotechnology Information GenBank
database to determine if a yet-undiscovered phage has been found.
- If the submission is a new phage, the discoverer has the chance to choose the bacteriophage’s
name.
- At the end of the spring semester, one ACM student and a faculty member present their
work at the annual National SEA-PHAGES Symposium. Students can also present their
results at the annual Maryland STEM Conference.
UMBC (University of Maryland, Baltimore County) is Allegany College of Maryland’s
buddy school in the SEA-PHAGES (Science Education Alliance-Phage Hunters Advancing
Genomics and Evolutionary Science) program. ACM became a SEA-PHAGES institution in 2019 and 59 students have been part of the
college’s PhageHunters program.
SEA-PHAGES program is a two-semester, discovery-based undergraduate research course
that that begins with students digging in the soil to find new viruses but progresses
through a variety of microbiology techniques and eventually to complex genome annotation
and bioinformatic analyses. It is jointly administered by the Howard Hughes Medical
Institute’s Science Education division and Graham Hatfull’s Lab within the Department
of Biological Sciences at the University of Pittsburgh.
More information about Allegany College of Maryland’s PhageHunters program is available at www.allegany.edu/phagehunters.







